Tackling Formatted Tabular Data from Excel
By Jeremy Selva
February 15, 2024
Overview
Reading data in formatted cell in excel can be really tricky. In this post, I will share six problematic formatted columns to read and share how I try to handle them.
Data Set
The data set can be found in this link:
Here is a peak view of the excel file with some problematic columns.
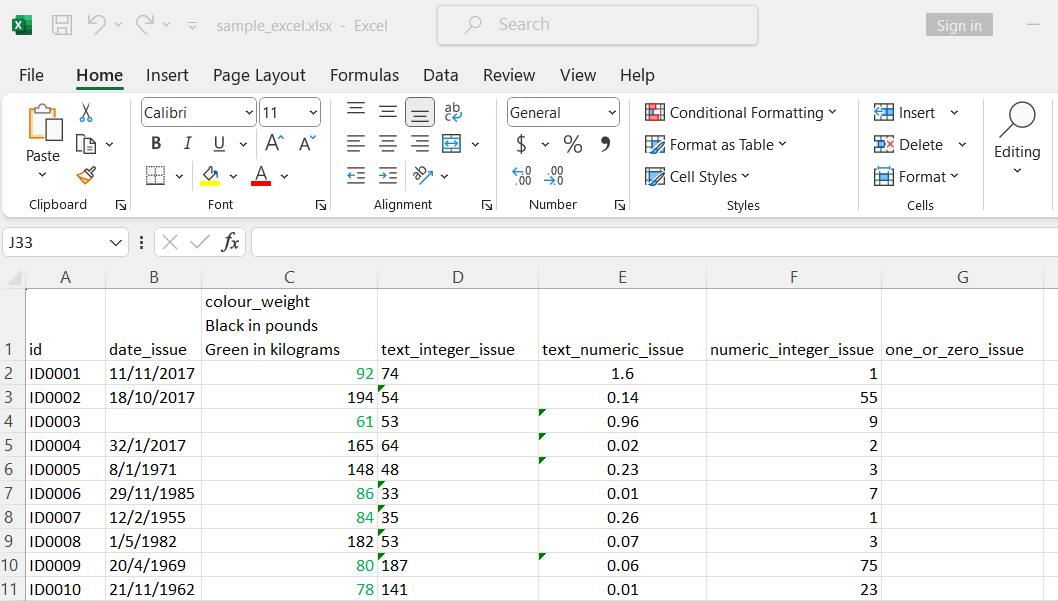
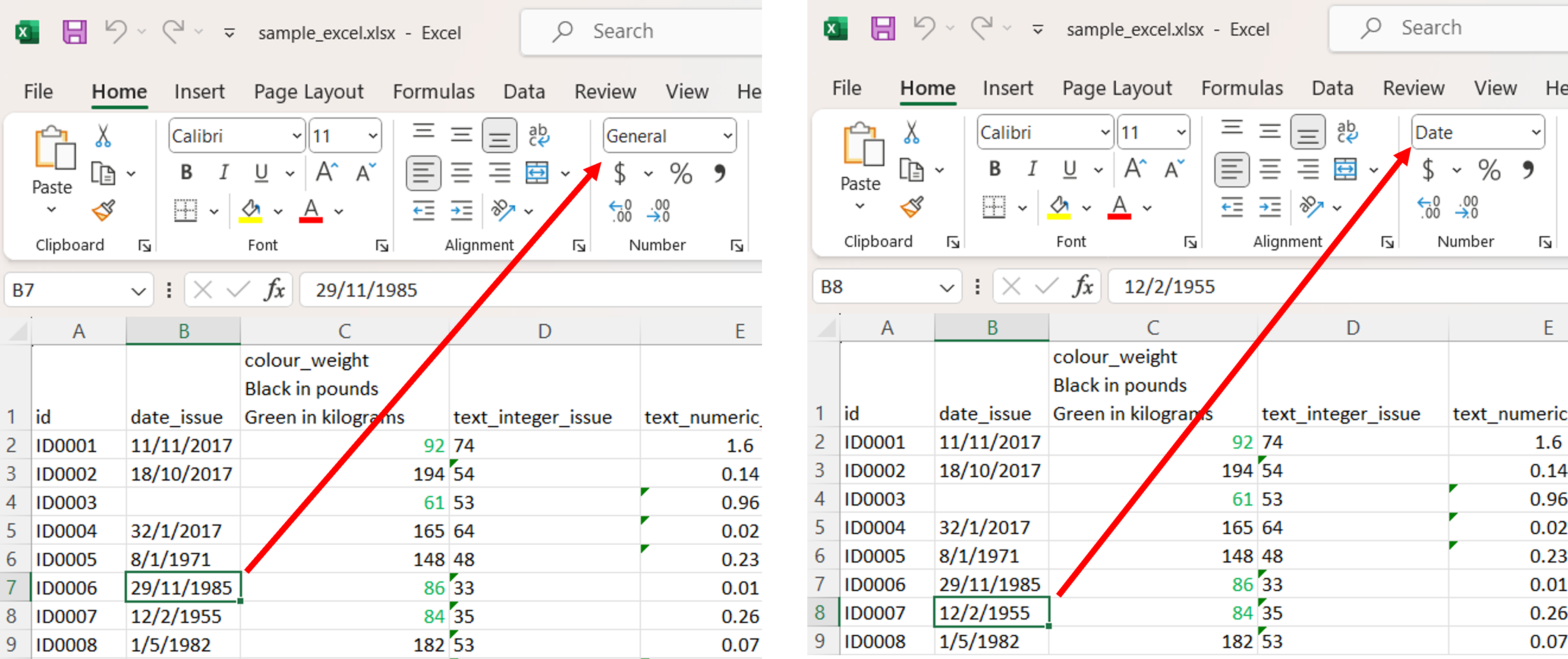
The column date_issue has two number format. One in General and the other in Date.
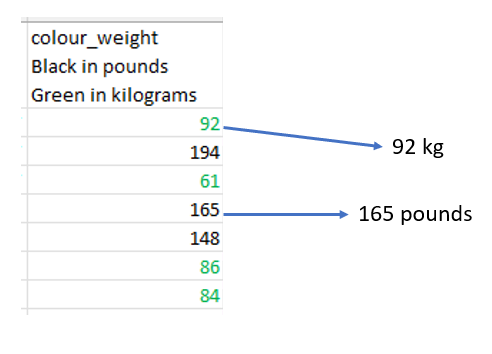
The column colour_weight has two colour format. The one in black is weight in pounds and the one in green is weight in kilogram.
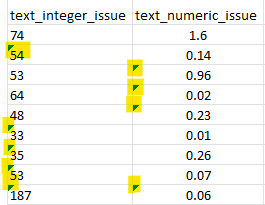
The columns text_integer_issue and text_numeric_issue are numeric columns but some cells were formatted as text. These cells are indicated by the green triangle.
The column numeric_integer_issue is an integer column.
The column one_or_zero_issue is a column containing one and zero but it is missing for the first hundreds of rows.
R Packages Used
Here are the R packages used in this analysis.
Code
library("sessioninfo")
library("quarto")
library("fs")
library("knitr")
library("here")
library("readxl")
library("collateral")
library("tidyxl")
library("janitor")
library("dplyr")
library("tidyr")
library("purrr")
library("lubridate")
library("tibble")
library("pointblank")
library("reactable")
library("unheadr")
Code
get_r_package_info <- function() {
r_package_table <- sessioninfo::package_info()
rownames(r_package_table) <- NULL
r_package_table <- r_package_table |>
tibble::as_tibble() |>
dplyr::mutate(
version = ifelse(is.na(r_package_table$loadedversion),
r_package_table$ondiskversion,
r_package_table$loadedversion)) |>
dplyr::filter(.data[["attached"]] == TRUE) |>
dplyr::select(
dplyr::any_of(c("package", "version",
"date", "source"))
)
return(r_package_table)
}
get_r_package_info() |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
R Platform Information
Here are the R platform environment used in this analysis.
Code
get_quarto_version <- function(
test_sys_path = FALSE,
test_no_path = FALSE
) {
# Taken from https://github.com/r-lib/sessioninfo/issues/75
if (isNamespaceLoaded("quarto") && isFALSE(test_sys_path)) {
path <- quarto::quarto_path() |>
fs::path_real()
ver <- system("quarto -V", intern = TRUE)
if (is.null(path) || isTRUE(test_no_path)) {
"NA (via quarto)"
} else {
paste0(ver, " @ ", path, "/ (via quarto)")
}
} else {
path <- Sys.which("quarto") |>
fs::path_real()
if (path == "" || isTRUE(test_no_path)) {
"NA"
} else {
ver <- system("quarto -V", intern = TRUE)
paste0(ver, " @ ", path)
}
}
}
get_knitr_version <- function() {
knitr_info <- "NA"
r_package_table <- sessioninfo::package_info(
pkgs = c("installed")
) |>
dplyr::filter(.data[["package"]] == "knitr")
if (nrow(r_package_table) == 1) {
knitr_version <- r_package_table$loadedversion[1]
knitr_source <- r_package_table$source[1]
knitr_info <- paste0(
knitr_version, " from ",
knitr_source)
}
return(knitr_info)
}
get_r_platform_info <- function() {
r_platform_table <- sessioninfo::platform_info()
r_platform_table[["quarto"]] <- get_quarto_version()[1]
r_platform_table[["knitr"]] <- get_knitr_version()[1]
r_platform_table <- data.frame(
setting = names(r_platform_table),
value = unlist(r_platform_table,
use.names = FALSE),
stringsAsFactors = FALSE
)
return(r_platform_table)
}
r_platform_table <- get_r_platform_info()
r_platform_table |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
Read Data Attempt 1
We try to read the formatted data but no warning was provided.
sample_excel_attempt_1 <- readxl::read_excel(
path = here::here("content",
"blog",
"2024-02-15-Tackling-Formatted-Cell-Data",
"sample_excel.xlsx"),
sheet = "Sheet1"
)
Though no warning was provided, when we print the variable, everything is turned to character and things are not read as expected.
date_issue: those formatted as Date have been turned to numberscolour_weight: different colour inputs not differentiatedtext_integer_issue: column turned to texttext_numeric_issue: column turned to textnumeric_integer_issue: column is read correctly as numericone_or_zero_issue: column turned to logical
Code
sample_excel_attempt_1 |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
Read Data Attempt 2
We try to read the formatted data indicating the right column types but it gives strange and intimidating warnings.
sample_excel_attempt_2 <- readxl::read_excel(
path = here::here("content",
"blog",
"2024-02-15-Tackling-Formatted-Cell-Data",
"sample_excel.xlsx"),
sheet = "Sheet1",
col_types = c("text" , "date",
"numeric", "numeric", "numeric", "numeric", "numeric")
)
This method resolves the issue when the one_or_zero_issue column being turned to logical by mistake.
Code
sample_excel_attempt_2 |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
On the other hand, the other issues are not fully resolved.
date_issue: those formatted as General have been turned to blankcolour_weightdifferent colour inputs not differentiated
From our previous failed attempt, it is also hard to convince ourselves and others that
- the numbers in characters from the columns
text_integer_issueandtext_numeric_issueare read correctly. - the numbers in
numeric_integer_issueonly have integer values. - the numbers in
one_or_zero_issueonly have values 0 or 1.
At this point of time, many of my colleagues actually gave up using R in their data analysis workflow because it is not working for them. For me, manually formatting these excel sheets does not work for me, especially when the data set is large.
There are limited resources to deal with formatted cells in Excel.
- https://nacnudus.github.io/spreadsheet-munging-strategies/tidy-formatted-cells.html
- unheadr::annotate_mf and unheadr::annotate_mf_all function from R package unheadr.
I will provide try to provide some ways to deal with these formatted columns mainly using collateral, pointblank, tidyxl.
Read Data Attempt 3
First, I convert the date columns into a list of character, Date and logical vectors so that I preserve both the text and date format.
sample_excel_attempt_3 <- readxl::read_excel(
path = here::here("content",
"blog",
"2024-02-15-Tackling-Formatted-Cell-Data",
"sample_excel.xlsx"),
sheet = "Sheet1",
col_types = c("text" , "list",
"text", "text", "text", "numeric", "numeric")
) |>
janitor::clean_names() |>
# Check if cohort id is unique
pointblank::rows_distinct(
columns = "id",
)
Code
sample_excel_attempt_3 |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
Fix date
Next I create a function that convert dates in character vectors into Date objects, convert logical vector to NA and convert Date vectors into the date format that I want.
convert_dmy_text_to_date <- function(input) {
if (length(class(input)) == 1) {
if (class(input) == "character") {
return(as.Date.character(lubridate::dmy(input)))
} else if (class(input) == "logical") {
return(NA)
}
}
return(as.Date.character(lubridate::ymd(input)))
}
However, creating function can lead to unexpected warnings and errors. To view these issues, I use some functions from the collateral R package. collateral::map_peacefully is able to capture various warnings and errors if any and summarised them as a tibble. collateral::has_warnings and collateral::has_errors returns a logical vector indicating which row has a warning.
To get the output is a little tricky, I actually have to the developer of collateral for help in this by opening a GitHub issue. For my case, the purrr::map_vec works for me.
fixed_date <- sample_excel_attempt_3 |>
dplyr::select(c("id","date_issue")) |>
dplyr::mutate(
converted_date_log = collateral::map_peacefully(
.x = .data[["date_issue"]],
.f = convert_dmy_text_to_date
),
converted_date = purrr::map_vec(
.x = .data[["converted_date_log"]],
.f = "result"
),
warning_check = collateral::has_warnings(.data[["converted_date_log"]]),
error_check = collateral::has_errors(.data[["converted_date_log"]]),
)
Code
fixed_date |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
I then use the function
pointblank::test_col_vals_in_set from the
pointblank package to detect if there are any issues by setting no_issue to be either TRUE or FALSE.
no_issue <- fixed_date |>
pointblank::test_col_vals_in_set(
columns = c("warning_check", "error_check"),
set = c(FALSE)
)
If there are any issues, I will isolate and display them.
if (!isTRUE(no_issue)) {
fixed_date |>
dplyr::filter(
warning_check == TRUE | error_check == TRUE
) |>
dplyr::mutate(
warning_log = purrr::map(.x = .data[["converted_date_log"]],
.f = "warnings",
.null = NA),
error_log = purrr::map(.x = .data[["converted_date_log"]],
.f = "errors",
.null = NA)
) |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
}
I will then correct the invalid dates accordingly and proceed. We just assume that the date was supposed to be 31-01-2017.
fixed_date <- sample_excel_attempt_3 |>
dplyr::select(c("id","date_issue")) |>
dplyr::mutate(
date_issue = dplyr::case_when(
(.data[["id"]] == "ID0004" &
.data[["date_issue"]] == "32/1/2017"
) ~ list(c("31/1/2017")),
.default = .data[["date_issue"]]
)
) |>
dplyr::mutate(
converted_date_log = collateral::map_peacefully(
.x = .data[["date_issue"]],
.f = convert_dmy_text_to_date
),
converted_date = purrr::map_vec(
.x = .data[["converted_date_log"]],
.f = "result"
),
warning_check = collateral::has_warnings(.data[["converted_date_log"]]),
error_check = collateral::has_errors(.data[["converted_date_log"]]),
) |>
pointblank::col_vals_in_set(
columns = c("warning_check", "error_check"),
set = c(FALSE)
) |>
dplyr::select(
c("id", "converted_date")
) |>
dplyr::rename(
date_fixed_yyyy_mm_dd = "converted_date"
)
Code
fixed_date |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
Fix weight (Method 1)
We use tidyxl function tidyxl::xlsx_cells to read the excel cell content (data values and some meta data of the cell).
cells <- tidyxl::xlsx_cells(
path = here::here("content",
"blog",
"2024-02-15-Tackling-Formatted-Cell-Data",
"sample_excel.xlsx"),
sheet = "Sheet1",
include_blank_cells = TRUE)
We observe that each cell is assigned to a local format id. We need to identify which local format id refers to the cell with black colour Calibri font and green colour Calibri font.
Code
cells |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
We use tidyxl function tidyxl::xlsx_formats to read the excel file in formats.
formats <- tidyxl::xlsx_formats(
path = here::here("content",
"blog",
"2024-02-15-Tackling-Formatted-Cell-Data",
"sample_excel.xlsx")
)
Using green as the running example, first in excel, click on a cell with green font. Next, click on the drop down button beside the font colour button.
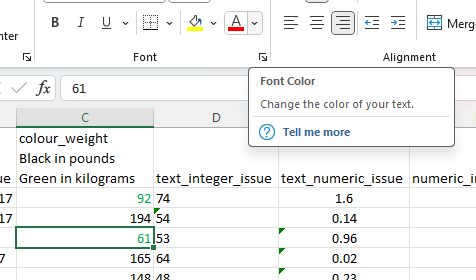
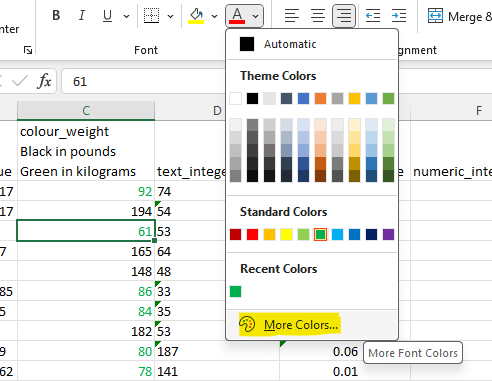
This will give the following output. Next click on More Colors…
Go to the Custom tab and extract the hex code saying #00B050 for green.
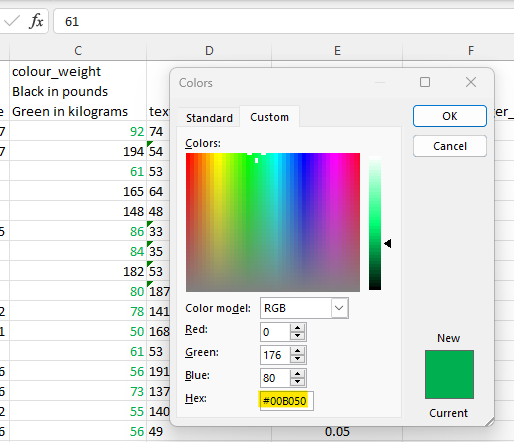
Next, use https://www.schemecolor.com/?getcolor={hex code} ( https://www.schemecolor.com/sample?getcolor=00B050 in our running example) to find out what the Hex8 code is for the green font.
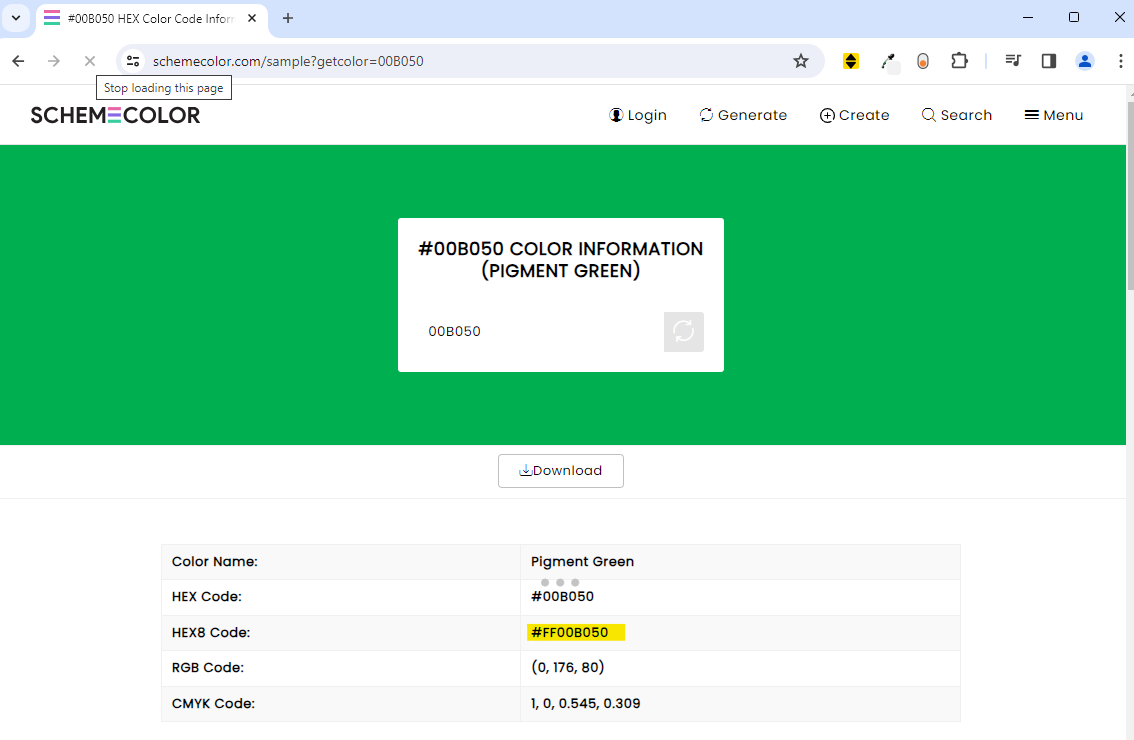
We can do a similar step to obtain the Hex8 code for the black font.
As such, we can identify the local format id accordingly with the black and green Hex8 code as #FF000000 and #FF00B050 respectively.
green_font_local_format_id <- which(formats$local$font$color$rgb == "FF00B050")
black_font_local_format_id <- which(formats$local$font$color$rgb == "FF000000")
green_font_local_format_id
[1] 3 14
black_font_local_format_id
[1] 1 2 4 5 6 7 8 9 10 15
We identify the column index of colour_weight_black_in_pounds_green_in_kilograms.
weight_column_index <- which(
colnames(sample_excel_attempt_3) == "colour_weight_black_in_pounds_green_in_kilograms"
)
weight_column_index
[1] 3
With the column index and local format id identified, we can filter the cells data to isolate cells which contain the weight in pounds and kilogram.
weight_pounds_converted <- cells |>
dplyr::filter(.data[["row"]] != 1) |>
dplyr::filter(.data[["col"]] == weight_column_index) |>
dplyr::filter(.data[["local_format_id"]] %in% black_font_local_format_id) |>
pointblank::col_vals_in_set(
columns = c("data_type"),
set = c("numeric")
) |>
dplyr::filter(.data[["data_type"]] == "numeric") |>
dplyr::select(c("row", "numeric")) |>
dplyr::rename(weight_pounds_converted = "numeric") |>
dplyr::mutate(
weight_pounds_converted =
janitor::round_half_up(.data[["weight_pounds_converted"]] / 2.2046,
digits = 0)
)
Code
weight_pounds_converted |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
weight_in_kg <- cells |>
dplyr::filter(.data[["row"]] != 1) |>
dplyr::filter(.data[["col"]] == weight_column_index) |>
dplyr::filter(.data[["local_format_id"]] %in% green_font_local_format_id) |>
pointblank::col_vals_in_set(
columns = c("data_type"),
set = c("numeric")
) |>
dplyr::filter(.data[["data_type"]] == "numeric") |>
dplyr::select(c("row", "numeric")) |>
dplyr::rename(weight_kg = "numeric")
Code
weight_in_kg |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
We can filter the cells data to isolate cells which contain the ids.
id_column_index <- which(
colnames(sample_excel_attempt_3) == "id"
)
id_cells <- cells |>
dplyr::filter(.data[["row"]] != 1) |>
dplyr::filter(.data[["col"]] == id_column_index) |>
pointblank::col_vals_in_set(
columns = c("data_type"),
set = c("character")
) |>
dplyr::select(c("row", "character")) |>
dplyr::rename(id = "character")
We combine the weight data together.
fixed_weight <- id_cells |>
dplyr::left_join(weight_pounds_converted,
by = dplyr::join_by("row"),
unmatched = "error",
relationship = "one-to-one") |>
dplyr::left_join(weight_in_kg,
by = dplyr::join_by("row"),
unmatched = "error",
relationship = "one-to-one") |>
tidyr::unite(
col = "weight_fixed_kg",
c("weight_pounds_converted",
"weight_kg"),
remove = TRUE,
na.rm = TRUE) |>
dplyr::select(c("id", "weight_fixed_kg"))
Code
fixed_weight |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)
Fix weight (Method 2)
An alternative method is to use
unheadr::annotate_mf_all from the
unheadr package. However the last column one_or_zero_issue must be removed.
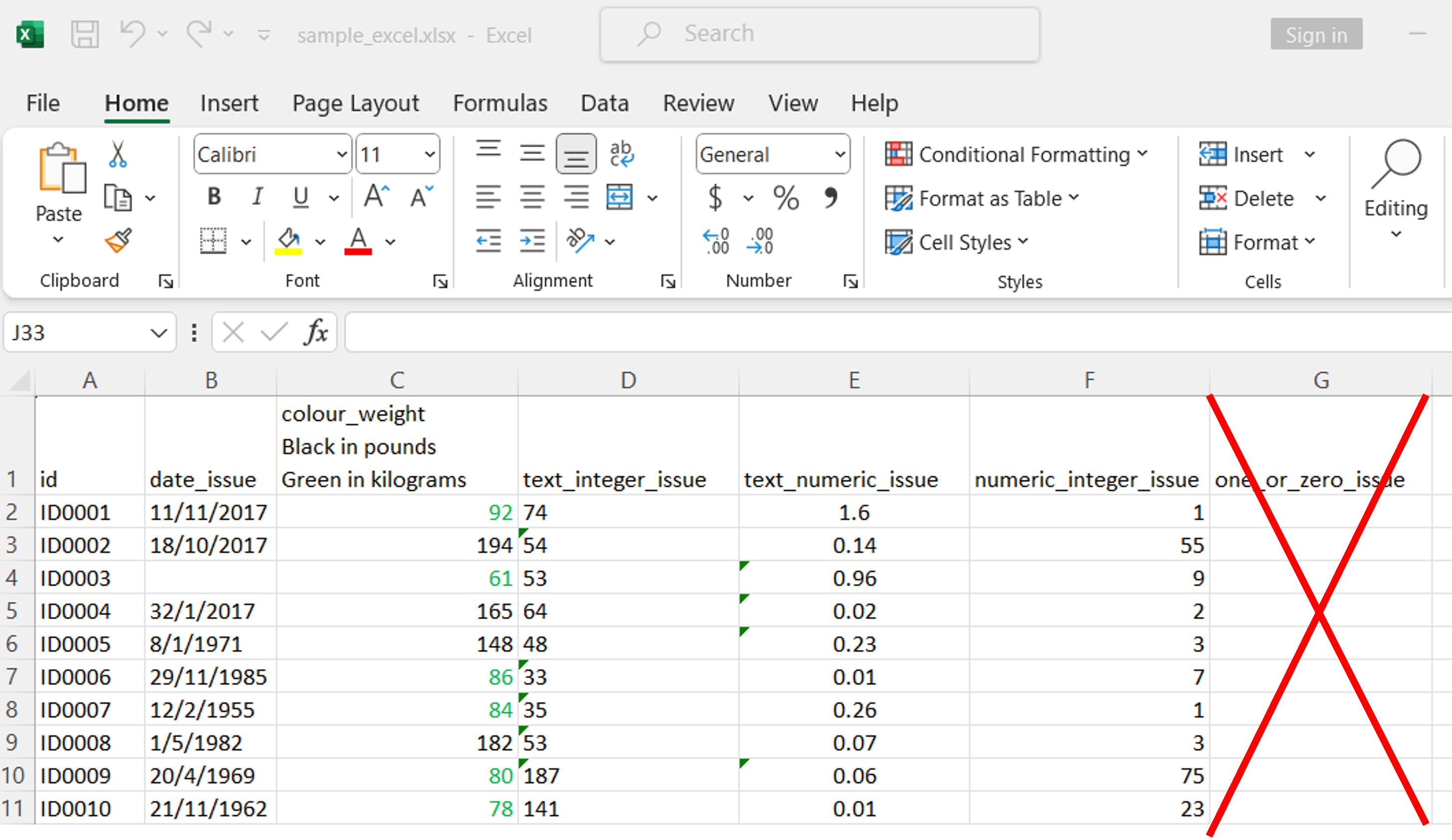
If not, the following error will be displayed
sample_excel_attempt_4 <- unheadr::annotate_mf_all(
xlfilepath = here::here("content",
"blog",
"2024-02-15-Tackling-Formatted-Cell-Data",
"sample_excel.xlsx")
)
Error in unheadr::annotate_mf_all(xlfilepath = here::here("content", "blog", : Check spreadsheet for blank cells in seemingly empty rows
Here is the file with the last column removed:
and the output of reading this file with unheadr::annotate_mf_all.
sample_excel_attempt_4 <- unheadr::annotate_mf_all(
xlfilepath = here::here("content",
"blog",
"2024-02-15-Tackling-Formatted-Cell-Data",
"sample_excel_remove_last.xlsx")
)
Code
sample_excel_attempt_4 |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump"
)
Check integer in character
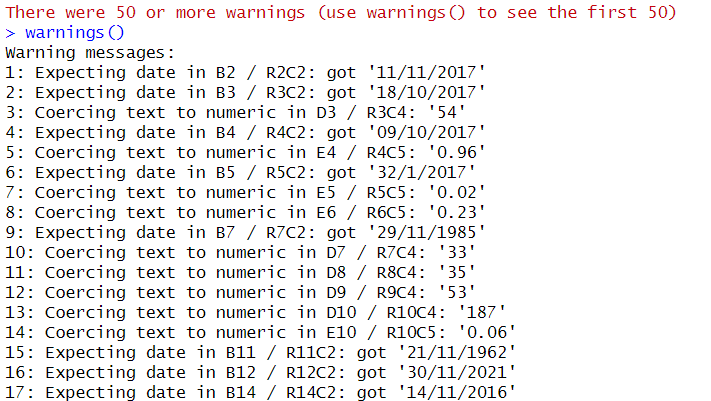
When we tried to read the excel data earlier (sample_excel_attempt_2) using col_types as numeric for the text_integer_issue column, we received a large chunk of error. Here are the relevant warning messages in relation to this column.
- Warning: Coercing text to numeric in D3 / R3C4: ‘54’
- Warning: Coercing text to numeric in D7 / R7C4: ‘33’
- Warning: Coercing text to numeric in D8 / R8C4: ‘35’
- Warning: Coercing text to numeric in D9 / R9C4: ‘53’
- Warning: Coercing text to numeric in D10 / R10C4: ‘187’
The warnings inform the user that it sees “54”, “33”, “35” and “53” in cells D3, D7, D8 and D9 respectively as “text” and it is forced to be converted to numeric. While in these cases, there are no issues when they are converted to numeric.
It may be safer to check if the column truly contain only positive integers even though they are in text, rather than relying on the long warning messages.
We continue with sample_excel_attempt_3 which reads the text_integer_issue column as text instead. We use some functions from the
pointblank package like
pointblank::col_vals_regex and
pointblank::col_vals_gt to ensure that the text are positive integers.
integer_check_from_text <- sample_excel_attempt_3 |>
dplyr::select(c("id","text_integer_issue")) |>
pointblank::col_vals_regex(
columns = c("text_integer_issue"),
regex = "^[1-9]([0-9]+)?(.[0]+)?$",
na_pass = TRUE,
) |>
dplyr::mutate(
text_integer_issue = as.integer(.data[["text_integer_issue"]])
) |>
pointblank::col_vals_gt(
columns = c("text_integer_issue"),
value = 0,
na_pass = TRUE,
) |>
dplyr::rename(
text_integer_fixed = "text_integer_issue"
)
Check numeric in character
When we tried to read the excel data earlier (sample_excel_attempt_2) using col_types as numeric for the text_integer_issue column, we received a large chunk of error. Here are the relevant warning messages in relation to this column.
- Warning: Coercing text to numeric in E4 / R4C5: ‘0.96’
- Warning: Coercing text to numeric in E5 / R5C5: ‘0.02’
- Warning: Coercing text to numeric in E6 / R6C5: ‘0.23’
- Warning: Coercing text to numeric in E10 / R10C5: ‘0.06’
The warnings inform the user that it sees “0.96”, “0.02”, “0.23” in cells E4, E5 and E6 respectively as “text” and it is forced to be converted to numeric.
Similarly, we can use the same functions pointblank::col_vals_regex and pointblank::col_vals_gt to ensure that the text are positive numbers. Do note that when numeric are read as text in this dataset. It can be of the form
- “0.140”
- “7.07E-2”
numeric_check <- sample_excel_attempt_3 |>
dplyr::select(c("id","text_numeric_issue")) |>
pointblank::col_vals_regex(
columns = c("text_numeric_issue"),
regex = "^[0-9]+((.[0-9]+)?(E(-)?[0-9]+)?)?$",
na_pass = TRUE,
) |>
dplyr::mutate(
text_numeric_issue = as.numeric(.data[["text_numeric_issue"]])
) |>
pointblank::col_vals_gt(
columns = c("text_numeric_issue"),
value = 0,
na_pass = TRUE,
) |>
dplyr::rename(
text_numeric_fixed = "text_numeric_issue"
)
Check integers in numeric
It may be necessary to check if a column only has integers. However, when using the function readxl::read_excel with col_types set to “numeric”. There is no way to check if the column truly contain integers, even when no warning is displayed. We also cannot use
pointblank::col_vals_regex because the column is not in text.
We create some functions to check if a numeric vector only has integers. The is_integer_vector returns FALSE when at least one of its element is not an integer.
is_integer_value <- function(input_value,
allow_na = FALSE) {
boolean_result <- FALSE
# When input value is NA
if (is.na(input_value)) {
if (isTRUE(allow_na)) {
boolean_result <- TRUE
return(boolean_result)
} else {
return(boolean_result)
}
}
# When input value is not numeric
if (isTRUE(!is.numeric(input_value))) {
return(boolean_result)
}
# When input value is numeric
boolean_result <- isTRUE(input_value %% 1 == 0)
return(boolean_result)
}
is_integer_vector <- function(input_vector,
allow_na = FALSE) {
boolean_results <- input_vector |>
purrr::map_lgl(
.f = is_integer_value,
allow_na = allow_na
)
return(boolean_results)
}
We use
pointblank::col_vals_expr to do a validation using our custom made is_integer_vector function.
Here is a case when it fails.
numeric_check |>
pointblank::expect_col_vals_expr(
expr = ~ is_integer_vector(numeric_check[["text_numeric_fixed"]],
allow_na = TRUE)
)
Error: The `expect_col_vals_expr()` validation failed beyond the absolute threshold level (1).
* failure level (1049) >= failure threshold (1)
We apply the is_integer_vector function on the numeric_integer_issue column before converting the column to an integer column.
integer_check_from_numeric <- sample_excel_attempt_3 |>
dplyr::select(c("id","numeric_integer_issue")) |>
pointblank::col_vals_expr(
expr = ~ is_integer_vector(sample_excel_attempt_3[["numeric_integer_issue"]],
allow_na = TRUE)
) |>
dplyr::mutate(
numeric_integer_issue = as.integer(.data[["numeric_integer_issue"]]),
) |>
dplyr::rename(
numeric_integer_fixed = "numeric_integer_issue"
)
Check values in set
For the column one_or_zero_issue, we use the function
pointblank::col_vals_in_set to check if the column contains 0, 1 or NA.
one_or_zero_check <- sample_excel_attempt_3 |>
dplyr::select(c("id","one_or_zero_issue")) |>
pointblank::col_vals_in_set(
columns = c("one_or_zero_issue"),
set = c(NA, 0, 1)
) |>
dplyr::rename(
one_or_zero_fixed = "one_or_zero_issue"
)
Combine
With the issues resolved for the problematic columns mentioned earlier, we combine them together to give our cleaned dataset.
cleaned_data <- sample_excel_attempt_3 |>
dplyr::select("id") |>
dplyr::left_join(fixed_date,
by = dplyr::join_by("id"),
unmatched = "error",
relationship = "one-to-one") |>
dplyr::left_join(fixed_weight,
by = dplyr::join_by("id"),
unmatched = "error",
relationship = "one-to-one") |>
dplyr::left_join(integer_check_from_text,
by = dplyr::join_by("id"),
unmatched = "error",
relationship = "one-to-one") |>
dplyr::left_join(numeric_check,
by = dplyr::join_by("id"),
unmatched = "error",
relationship = "one-to-one") |>
dplyr::left_join(integer_check_from_numeric,
by = dplyr::join_by("id"),
unmatched = "error",
relationship = "one-to-one") |>
dplyr::left_join(one_or_zero_check,
by = dplyr::join_by("id"),
unmatched = "error",
relationship = "one-to-one")
Code
cleaned_data |>
reactable::reactable(
filterable = TRUE,
defaultPageSize = 5,
paginationType = "jump",
)